Navigation auf uzh.ch
Navigation auf uzh.ch
Sensing the chemical environment is essential to all life forms, and the specific detection of volatile chemostimuli ensures rapid and appropriate responses in various animal species. Unlike animals, plants do not possess olfactory receptors neither associated neural networks. However, plant cells do communicate via volatile organic compounds (VOCs) such as benzenoids, terpenoids and C6-aldehydes. The key defense-related phytohormones ethylene, methyl jasmonate and methyl salicylate are bona fide volatiles that trigger profound reprogramming of the plant transcriptome into an optimized growth/immune response trade-off.
Our research questions the role of bacteria-derived VOCs as signaling cues between microbes and plants. Bacterial VOCs bouquets comprise plant-like and microbe-specific molecules that elicit plant growth and systemic resistance. Yet, it remains unclear how plants perceive such molecules and distinguish bacterial cues from plant signals. The overlap between plant and microbial volatilomes suggests that VOCs constitute an ancient cross-kingdom language that allows the early recognition of compatible or incompatible partnerships.
We focus on the VOCs-based interaction between Arabidopsis thaliana and rhizospheric Pseudomonas and Burkholderia species. Both bacterial genera display plant health-promoting and phytopathogenic strains. Their biogenic emissions (volatilomes), rich of about 200 identified compounds mostly issued from basic metabolism, elicit A. thaliana systemic resistance and enhance its secondary root architecture. Pseudomonas and Burkholderia share about 20% of these VOCs. Therefore, a large number of volatile species could represent genus- or strain-specific chemical signatures perceived by plants. Since a constant mounting of the defense status would be a tremendous cost for plant growth and other physiological functions, we currently question the specificity of this mechanism and its ecological role. To understand if plants preferentially perceive self-like molecules or scan for genuine bacterial cues, we strive to identify the underlying molecular pathways using well-established Arabidopsis toolsets.
Funding: Swiss National Science Foundation (SNF), Sinergia grant CRSII3_154430
An obligate plant-bacterium symbiosis was described between the tropical plant Psychotria kirkii and its bacterial symbiont Candidatus Burkholderia kirkii. The symbiosis is phenotypically characterised by the presence of leaf nodules filled with bacteria. (Figure 1). The plant hosts a reservoir of undifferentiated bacterial cells at its apical meristems, which invade the stomatal chamber of developing leaves and eventually become pleomorphic in the mature nodule. The bacteria are found throughout plant development and are vertically transmitted via the seeds. Despite the fact that a century of research has passed, the principles governing this strictly dependent relationship it is largely unknown. This lack of knowledge is largely due to the fact that the plant and its symbiont cannot be grown outside of the symbiosis. Total genome sequencing analysis of the bacterial symbiont showed evidence of massive genome erosion with many non-essential genes lost through this process, but unexpectedly conserved genes related to secondary metabolism. We isolated and characterized two secondary metabolites of bacterial origin, namely kirkamide and streptol glucoside. Kirkamide belongs to the family of C7N aminocyclitols and has been found to be insecticidal while the conjugated cyclitol streptol glucoside seems to be a potent allelochemical.
A multidisciplinary approach joining the expertise in microbiology (Prof. Eberl’s group), chemistry (Prof. Gademann’s group, UZH) and plant biology (Thomas Wicker’s group, UZH) within the frame of this Sinergia project, aims at understanding the molecular basis underlying this unique symbiosis.

Figure 1 Psychotria kirkii nodulated leaf.
People involved: Antri Georgiou (PhD Candidate) and Dr. Aurélien Bailly
Collaborators: Dr. Hsiao Chien-Chi (Prof. Gademann’s group, UZH)
Streptol glucoside (SG), a microbial secondary metabolite produced from the nodulated P. kirkii leaves, was found to be a potent allelopathic compound that prevented the germination of a broad range of plant species, but not of P. kirkii.
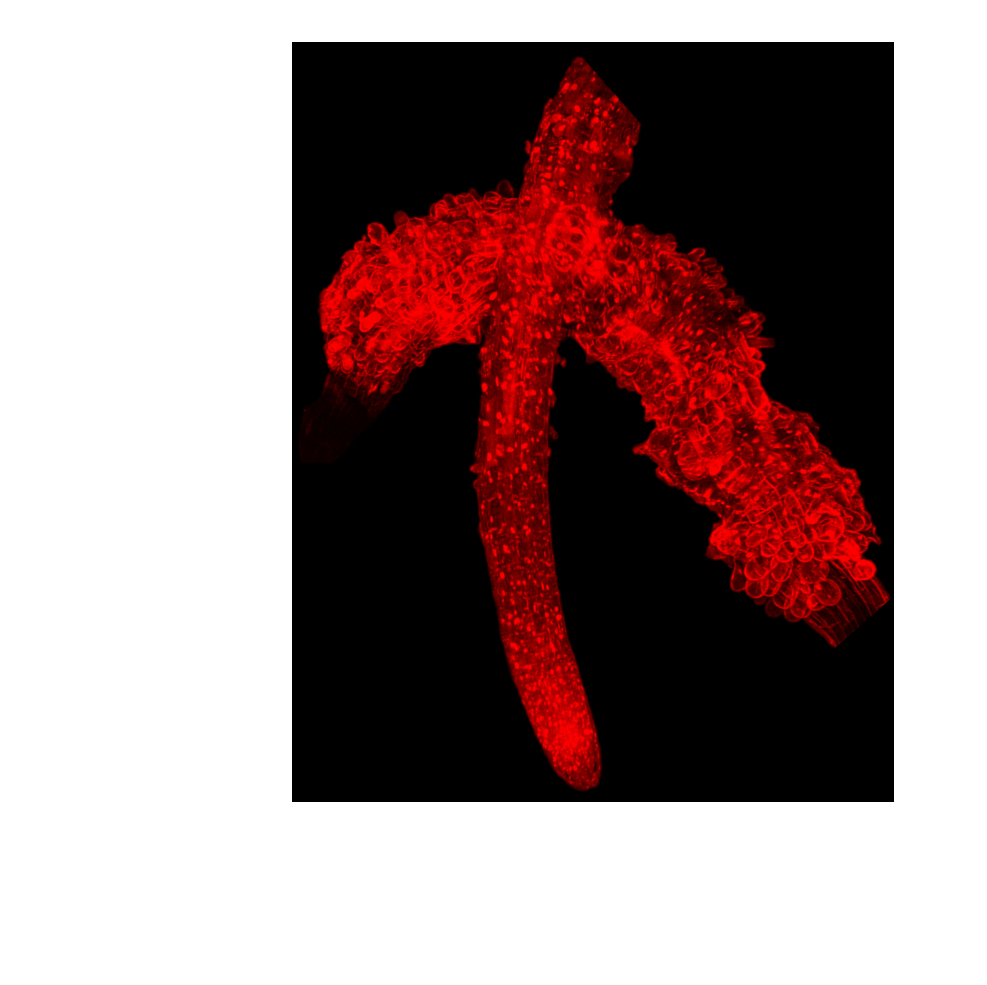
Our research goal is to identify the potential biological role and targets of this compound. We are currently evaluating the activity of crude P. kirkii leaf extracts as well as the activity of pure, chemically synthetized streptol glucoside and associated derivatives in bioassays using the model plant Arabidopsis thaliana. The dramatic phenotypic abnormalities and developmental arrests caused by streptol glucoside treatments suggest a root-specific mode of action targeting cell wall integrity and remodelling. (Figure 2). Our ongoing work attempts to combine biochemical, transcriptomics and mutants screening approaches in A. thaliana in order to pinpoint the molecular targets of this unusual sugar-like compound.
Beyond understanding the fundamental role of Burkholderia secondary metabolites in plant-microbe interactions and their involvement in the establishment and maintenance of this very symbiosis, our work sets the basis to pharmaceutical lead designs of novel antidiabetic and antiviral molecules to promote and sustain human health.